Sleeper cells – the secret lives of invincible bacteria
Our antibiotics are failing us. Bacteria are evolving resistance to virtually all of our frontline drugs. They build up mutations in their DNA that allow them to pump drugs from their cells, or build enzymes that destroy them. They shift into stealth mode, by changing the shape of the molecules that antibiotics are designed to target.
This is the standard story of how microbes foil antibiotics, and it’s incomplete. Some bacteria can withstand an antibiotic assault without any special defences. They just keep their heads down.
All of our antibiotics have been designed to kill bacteria that are actively multiplying, like jamming a spanner into a whirring machine. If the machine is still, if no wheels or cogs are turning, the spanner does nothing. So by entering a dormant state, when they either grow extremely slowly or not at all, bacteria can withstand huge doses of antibiotics. They’re ‘tolerant’ rather than classically resistant. And later, these sleeper cells can awaken to start a new wave of infection, like a field of grass rebounding after being sliced by a mower.
These bacteria are called persisters. That is, after all, what they do – they survive, they endure. Persisters aren’t a particular species of bacteria – they’re more like a temporary occupation. They’re a state that microbes can switch to when threatened by antibiotics. Their ability to survive and rebound could help to explain why many infections, such as tuberculosis, take so long to treat.
But even this story is too simple. Yuichi Wakamoto and Neeraj Dhar from the Swiss Federal Institute of Technology have questioned the longstanding assumption that persisters don’t grow, or grow very slowly. By studying Mycobacterium smegmatis, a relative of the bug that causes tuberculosis, they found the opposite: Persisters do grow, but they die at an equal rate, creating the illusion of a static population.
They describe this strategy as “dynamic persistence”, and it’s a new one. That’s not to say that all persisters do this, but it shows just how little we know about the secret lives of bacteria. “It’s a real advance,” says Bruce Levin from Emory University. “It shows how little we know about how antibiotics do their killing.”
Wakamoto and Dhar’s ace card was a technique called microfluidics, which channels microbes along tiny pathways and allows scientists to track them individually. Rather than counting a swirling mass of bacteria growing (or not) in a flask, Wakamoto and Dhar could follow the fate of each cell—an indispensable technique for working out what they’re actually doing.
The duo exposed their microbes to isoniazid, an anti-tuberculosis drug that stops bacteria from building their outer walls. Isoniazid killed off almost 90 percent of the bacteria. The rest were persisters, but contrary to expectations, these survivors kept on growing through the drug assault. And slow-growing cells were just as likely to persist as fast-growing ones.
The graph below shows how the descendants of a single bacterium fared. It’s a family tree going from left to right, with each fork representing a moment when one cell split into two. The addition of isoniazid (the first dotted line) kills most of the cells, but a small number survive and, more importantly, keep on dividing until the drug is withdrawn (the second dotted line).
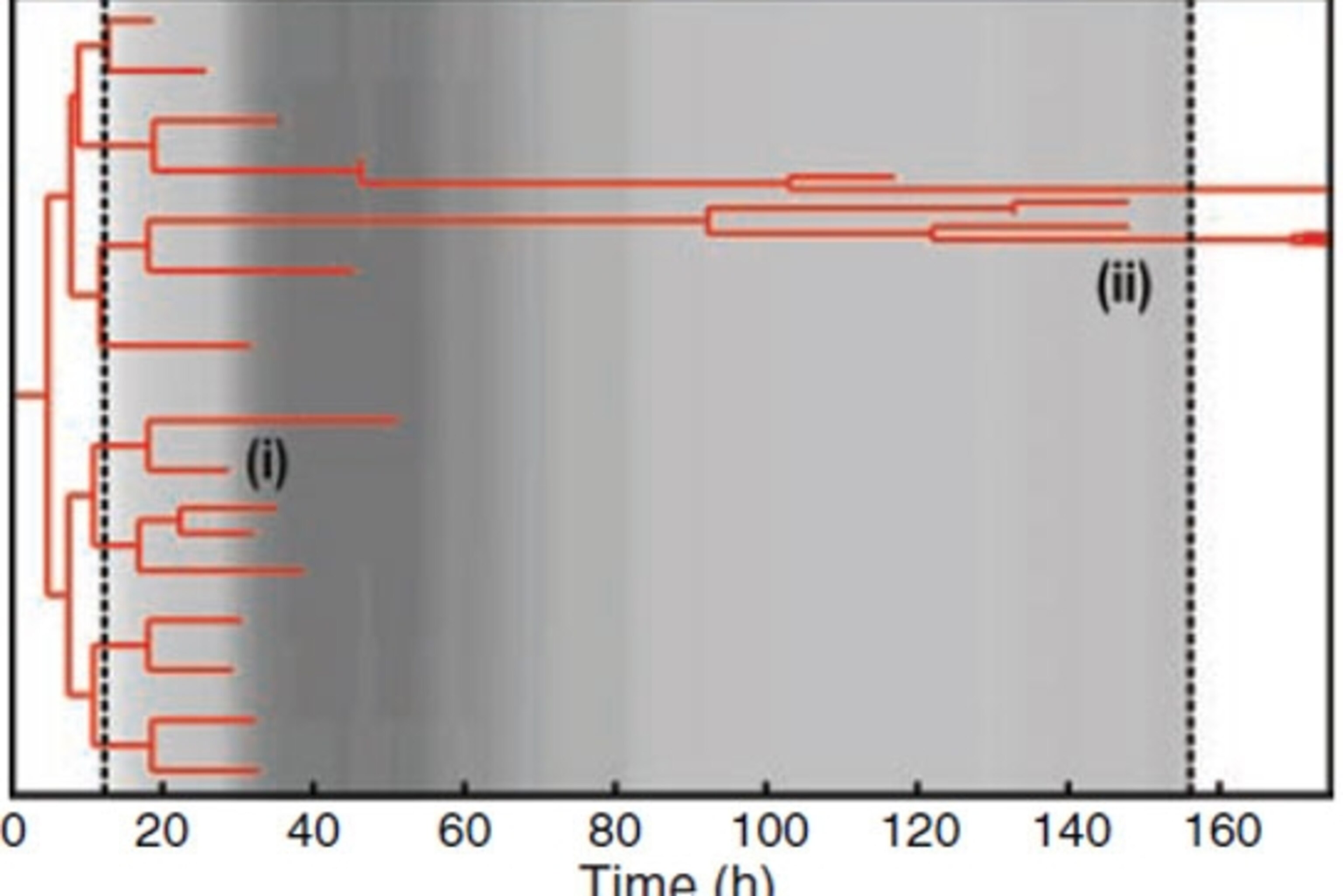
The birth and death rates of these bacteria were so finely balanced that to a careless eye, it looked like the population stayed the same. Only Wakamoto and Dhar’s microfluidics technique revealed the hidden cycle so life and death.
The persisters survive because isoniazid actually needs bacteria to sign their own death warrants. It only becomes active after being processed by an essential bacterial enzyme called KatG. It’s like handing someone a bomb in the knowledge that they will inevitably light the fuse.
But KatG isn’t always around. Bacteria manufacture the enzyme in random pulses and it’s the timing of these pulses that determines whether they survive a brush with isoniazid. If the drug hits a bacterium during the window when it’s making KatG, it becomes active and kills the cell. If it hits during a trough, there’s barely any KatG around, it stays inactive, and the bacteria persist.
This has important implications. While persisters aren’t classically resistant to antibiotics, they do increase the odds that such resistance will evolve. By enduring, they greatly increase the length of an infection and provides more time for each new generation of microbes to accrue adaptations against a continual wave of drugs. In Wakamoto and Dhar’s scenario, this threat looms even greater. If the cells can continue growing during an antibiotic blitz, rather than just after it, they face even greater evolutionary pressures to evolve resistance, and would be more likely to do so.
Naturally, this does not mean that all persisters behave in this way. Indeed, Kim Lewis from Northeastern University, one of the leaders in this field, is unsure whether Wakamoto and Dhar’s cells should be called persisters at all. “An important feature of persisters is that these cells are tolerant to many, essentially all drugs,” he says. They do this in diverse ways, by shutting down the machinery that drugs are designed to jam and destroy. That’s very different to what Wakamoto and Dhar found, in which bacteria resist one specific drug because of random cycles in one enzyme that activates it. “Dynamically resistant cells would be a better term,” says Lewis.
In some ways, it’s a semantic argument, and persister research is full of semantic arguments. These cells are also called tolerant, latent, indifferent, dormant and non-multiplying, which all have slightly different connotations. This may be because persisters themselves are so varied.
As Levin says, “The paper supports my prejudice that persistence is not a single phenomenon but many different phenomena.” This fits with the results of genetic studies. You can knock out virtually any single gene in bacteria like E.coli, and the resulting mutants will still form persisters. There are many ways of making a persister—a fact that undoubtedly contributes to the success of this strategy (and of bacteria in general) but also makes them fiendishly hard to study.
If you want to find out more about persisters, I wrote a feature about them for New Scientist last year. Download it here.
Reference: Wakamoto, Dhar, Chait, Schneider, Signorino-Gelo, Leibler & McKinney. 2013. Dynamic Persistence of Antibiotic-Stressed Mycobacteria. Science http://dx.doi.org/10.1126/science.1229858
More on drug resistance:
- Harmless soil bacteria are trading weapons with those that kill us
- Antibiotics fuel obesity by creating microbe upheavals
- Genome detectives unravel spread of stealthy bacteria in a hospital
- Isolated for millions of years, cave bacteria resist modern antibiotics
- Bacteria: resisting antibiotics since at least 30,000 BC
- Fighting evolution with evolution – using viruses to target drug-resistant bacteria
- Infectious bacteria in your gut create black market for weapons
- Scientists track the evolution of an epidemic to show how bacteria adapt
Go Further
Animals
- Octopuses have a lot of secrets. Can you guess 8 of them?
- Animals
- Feature
Octopuses have a lot of secrets. Can you guess 8 of them? - This biologist and her rescue dog help protect bears in the AndesThis biologist and her rescue dog help protect bears in the Andes
- An octopus invited this writer into her tank—and her secret worldAn octopus invited this writer into her tank—and her secret world
- Peace-loving bonobos are more aggressive than we thoughtPeace-loving bonobos are more aggressive than we thought
Environment
- Listen to 30 years of climate change transformed into haunting musicListen to 30 years of climate change transformed into haunting music
- This ancient society tried to stop El Niño—with child sacrificeThis ancient society tried to stop El Niño—with child sacrifice
- U.S. plans to clean its drinking water. What does that mean?U.S. plans to clean its drinking water. What does that mean?
- Food systems: supporting the triangle of food security, Video Story
- Paid Content
Food systems: supporting the triangle of food security - Will we ever solve the mystery of the Mima mounds?Will we ever solve the mystery of the Mima mounds?
History & Culture
- Strange clues in a Maya temple reveal a fiery political dramaStrange clues in a Maya temple reveal a fiery political drama
- How technology is revealing secrets in these ancient scrollsHow technology is revealing secrets in these ancient scrolls
- Pilgrimages aren’t just spiritual anymore. They’re a workout.Pilgrimages aren’t just spiritual anymore. They’re a workout.
- This ancient society tried to stop El Niño—with child sacrificeThis ancient society tried to stop El Niño—with child sacrifice
- This ancient cure was just revived in a lab. Does it work?This ancient cure was just revived in a lab. Does it work?
Science
- The unexpected health benefits of Ozempic and MounjaroThe unexpected health benefits of Ozempic and Mounjaro
- Do you have an inner monologue? Here’s what it reveals about you.Do you have an inner monologue? Here’s what it reveals about you.
- Jupiter’s volcanic moon Io has been erupting for billions of yearsJupiter’s volcanic moon Io has been erupting for billions of years
- This 80-foot-long sea monster was the killer whale of its timeThis 80-foot-long sea monster was the killer whale of its time
Travel
- How to plan an epic summer trip to a national parkHow to plan an epic summer trip to a national park
- This town is the Alps' first European Capital of CultureThis town is the Alps' first European Capital of Culture
- This royal city lies in the shadow of Kuala LumpurThis royal city lies in the shadow of Kuala Lumpur
- This author tells the story of crypto-trading Mongolian nomadsThis author tells the story of crypto-trading Mongolian nomads